Novembre Lab
Research and Training in Population Genetic Methods, Theory, and Application
Our research group develops and applies computational tools to study genetic diversity in natural populations. Our goal is to analyze and interpret large-scale genomic data to improve the understanding of: (1) basic genomic biology, (2) the biology of heritable disease traits, (3) the genetic basis of evolutionary processes, and (4) the history and evolution of various species, especially humans.
From a disciplinary perspective, most of the ideas we use are from theoretical population genetics, statistical genetics, and computational statistics. We enjoy analyzing data from emerging genotyping/sequencing technologies especially from large or particularly interesting population samples.
Research
Population Genetics Methods and Theory
Our core projects address the following broad problems:
-
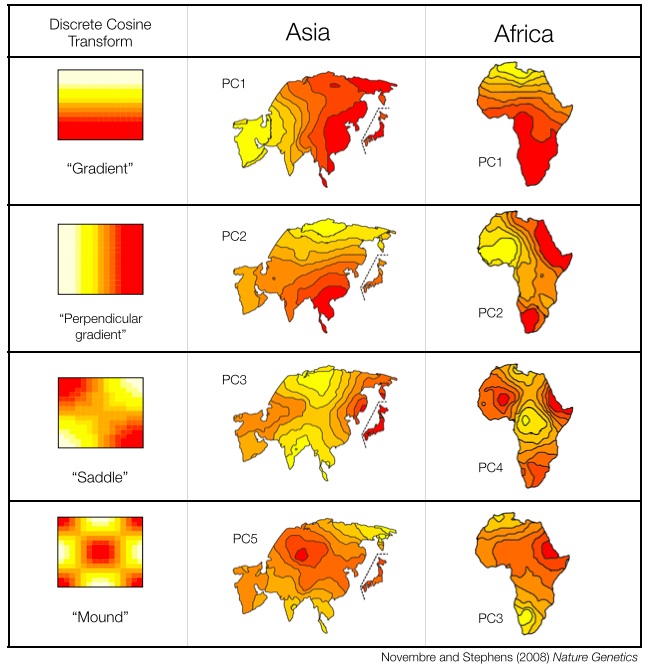
Methods for studying and visualizing population structure
-
Methods for leveraging haplotype data for methods for inference in population genetics
-
The impact of recent demographic processes, particularly rapid recent growth
-
Variation in the intensity of mutation and negative selection across the genomic landscape
Human Population Genetics
Much of our work focuses on human genetic data, motivated in equal parts by an inherent curiosity, by the biomedical importance, and by the relevance for a broader understanding of evolution in all organisms. Our projects address:
-
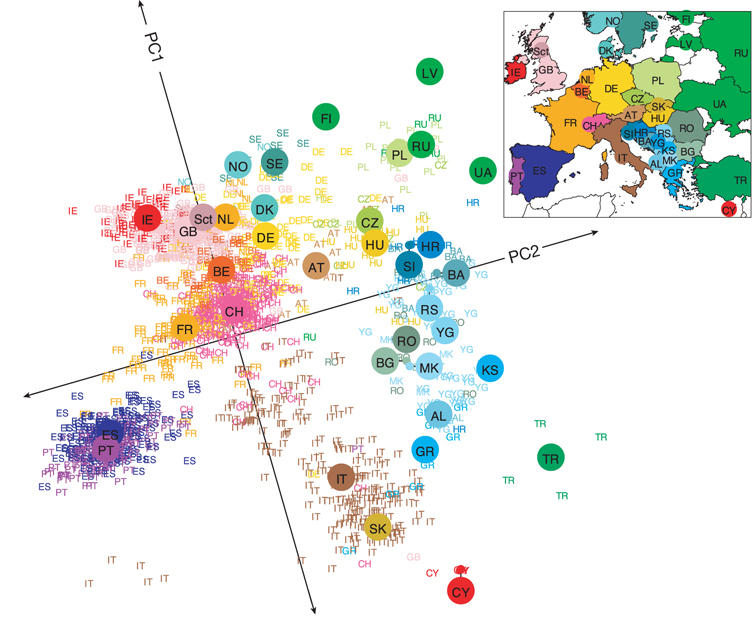
Patterns of population structure in human populations especially within world regions and finer spatial-scales
-
The impact of recent demographic processes, particularly rapid recent growth, on human genetics
-
The interaction of selection and demographic history in human evolutionary history
-
Inference of relative recombination rates from local ancestry patterns in admixed populations
-
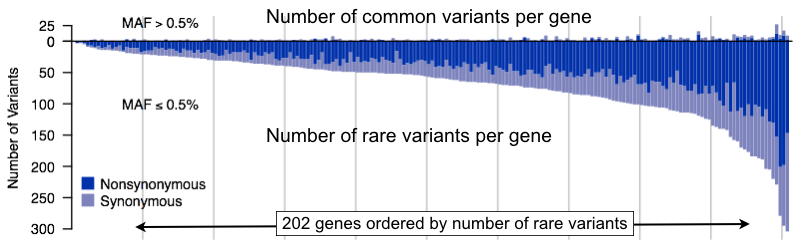
Correcting for population structure in human genome-wide association studies
-
Personalized genomics: Inference of detailed individual ancestry from genetic data
-
Human ancient DNA studies
Broader Evolutionary Genetics and Molecular Evolution
With the help of great collaborators, we have worked on several additional questions in evolutionary genetics and molecular evolution. Some past examples include:

-
Canid evolution: With Bob Wayne (UCLA), we work on problems regarding wolf population structure, early dog domestication, wolf-dog introgression, and canid mutation rates.
-
Dobzhansky-Muller incompabilitites (DMIs) in mice: With Bret Payseur (Wisconsin), we work on the local ancestry signatures made by DMIs.
-
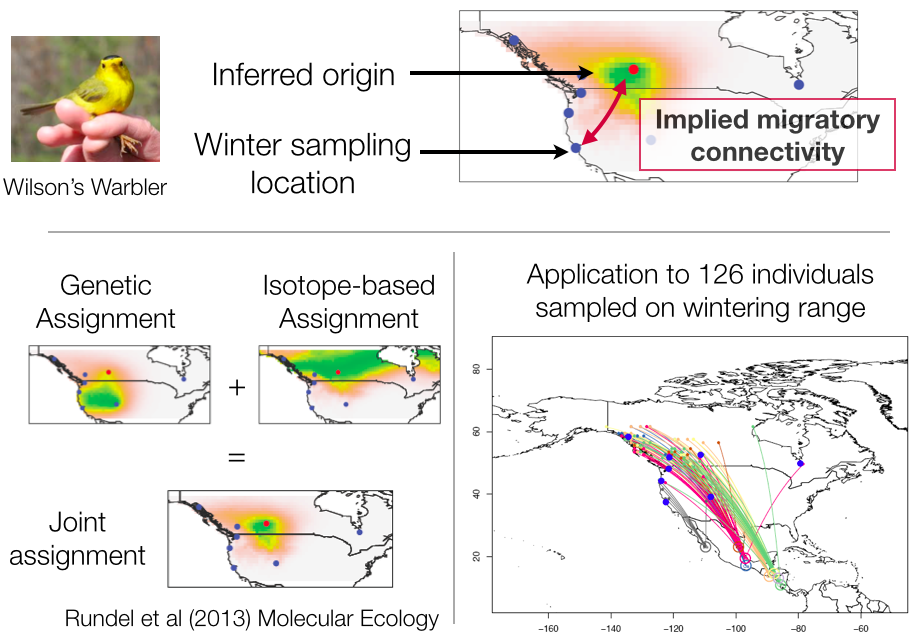
Avian migration: With Kristen Ruegg (UCSC) and Tom Smith (UCLA), we have worked on methods for studying seasonal bird migrations.
-
Evolve-and-resequence studies: With Tom Turner (UCSB), we have developed methods and simulations for understanding evolve-and-resequence studies in Drosophila.
-
Codon usage bias: With Dennis Wall and Josh Herbeck (when we were all back at UC-Berkeley), we produced a method for measuring for codon usage bias while accounting for background nucleotide composition.
Training
Beyond our research goals, we have the following training and outreach aims:
-
Train graduate students and postdoctoral fellows in the interdisciplinary skills necessary to make independent, insightful contributions to contemporary research problems in human genetics and evolutionary biology.
-
Develop teaching materials and courses to help: (1) prepare a new generation of biologists that will need strong skills in quantitative reasoning and computation to address the vast amounts of data now readily available; (2) prepare future citizens who are well-informed and capable of sound decision-making in a world in which science and particularly genetics will play an increasingly important role for society.
-
Promote international collaborative experiences for students to help foster individual growth and to strengthen scientific institutions and training programs through exchange.

John Novembre, PhD
Professor
Department of Human Genetics
Department of Ecology & Evolution
University of Chicago
BA, The Colorado College (2000)
PhD, University of California-Berkeley (2006)
email: jnovembre uchicago edu
twitter: @jnovembre
phone: 773-834-8217
office: Cummings 421
address: 920 E 58th Street, Chicago, IL 60636
[UChicago Profile]
[Curriculum vitae]
News
7/25Luke Anderson-Trocme leaves the lab to start his own group at the Université de Montréal!
6/25Maggie Steiner finished her PhD and starts a job at BCG in Chicago!
2/25Our paper building on EEMs/FEEMs to detect estimate asymmetric gene flow is now up on bioRxiv.
5/25Vivaswat Shastry finished his PhD and continues on in the Berg Lab (UChicago)!
2/25Our paper building on EEMs/FEEMs to detect evidence of long-range migration is now up on bioRxiv.
2/25Xinyi Li finished her PhD and joins the Pritchard Lab (Stanford)!
12/24Our paper on the effects of study design on the discovery of rare, deleterious variants is now up on bioRxiv! For a summary, see the associated Bluesky thread.
9/24Carla Hoge has joined the lab!
3/24Sherif won an honorable mention for his poster presentation at PEQG in Washington, DC.
3/24Carla Hoge has recieved a Chicago Fellows postdoctoral fellowship to join the lab later in 2024!